Andrew Dalke (talk | contribs) (Added image) |
Andrew Dalke (talk | contribs) (shortened the code so there's no scrollbar to display it) |
||
| Line 14: | Line 14: | ||
==OpenEye/Python== |
==OpenEye/Python== |
||
| − | [[ |
+ | [[Image:Align_oe.gif|thumb|right|alt=Aligned depictions with the OpenEye toolkits.|Output from OpenEye/Python]] |
| + | |||
| + | |||
<source lang="python"> |
<source lang="python"> |
||
from openeye.oechem import * |
from openeye.oechem import * |
||
| Line 26: | Line 28: | ||
view.SetAromaticCircles(False) |
view.SetAromaticCircles(False) |
||
view.SetAromaticDashes(True) |
view.SetAromaticDashes(True) |
||
| − | |||
view.SetColorOnBlack(True) |
view.SetColorOnBlack(True) |
||
view.SetBackColor(0, 0, 0) |
view.SetBackColor(0, 0, 0) |
||
view.SetForeColor(255, 255, 255) |
view.SetForeColor(255, 255, 255) |
||
| − | |||
view.SetShowHydrogens(True) |
view.SetShowHydrogens(True) |
||
view.SetDativeBonds(False) |
view.SetDativeBonds(False) |
||
| − | |||
return view |
return view |
||
| − | |||
# This assumes the input was in proper SD format already |
# This assumes the input was in proper SD format already |
||
| Line 44: | Line 42: | ||
OE3DToAtomStereo(mol) |
OE3DToAtomStereo(mol) |
||
OEAddDepictionHydrogens(mol) |
OEAddDepictionHydrogens(mol) |
||
| − | |||
core_subsearch = OESubSearch("[#7]~1~[#6]~[#6]~[#7]~[#6]~[#6]~2~[#6]~[#6]~[#6]~[#6]~[#6]12") |
core_subsearch = OESubSearch("[#7]~1~[#6]~[#6]~[#7]~[#6]~[#6]~2~[#6]~[#6]~[#6]~[#6]~[#6]12") |
||
| − | |||
ifs = oemolistream("benzodiazepine.sdf.gz") |
ifs = oemolistream("benzodiazepine.sdf.gz") |
||
| − | |||
img = OE8BitImage(CELL_WIDTH*4, CELL_HEIGHT*4) |
img = OE8BitImage(CELL_WIDTH*4, CELL_HEIGHT*4) |
||
view = make_view() |
view = make_view() |
||
| Line 60: | Line 55: | ||
if at_first_structure: |
if at_first_structure: |
||
OEDepictCoordinates(mol) |
OEDepictCoordinates(mol) |
||
| + | # save the first structure's depiction coordinates |
||
coords = [mol.GetCoords(atom) for atom in match.GetTargetAtoms()] |
coords = [mol.GetCoords(atom) for atom in match.GetTargetAtoms()] |
||
else: |
else: |
||
| + | # Align to the first structure's coordinates |
||
fixed = [0] * mol.GetMaxAtomIdx() |
fixed = [0] * mol.GetMaxAtomIdx() |
||
for atom, coord in zip(match.GetTargetAtoms(), coords): |
for atom, coord in zip(match.GetTargetAtoms(), coords): |
||
| Line 75: | Line 72: | ||
# Only align the first match |
# Only align the first match |
||
break |
break |
||
| − | else: |
||
| − | raise AssertionError("No match") |
||
# Stop after 16 structures |
# Stop after 16 structures |
||
Revision as of 00:30, 4 February 2010
This task comes from working with researchers who have a desktop tool which displays a large number (a few hundred) structures on a scrolling page. The structures are the database hits from searching with a given SMARTS query, and the common SMARTS substructures are aligned in each depictions. This makes it easier for them to identify the differences between the structures.
They want to replace it with something newer, and they would like more control over how the structure is displayed. (For example, some people want the depiction tool to first follow corporate guidelines on depiction layout. Doing that is outside the scope of this task.)
Implementation
Use the first 16 structures of the benzodiazepine SD file to make a 4x4 grid of depictions as a single image. The first structure is in the upper-left corner, the second is to its right, and so on. Each depiction should include the title field of the corresponding record, which in this case is the PubChem identifier.
Use "[#7]~1~[#6]~[#6]~[#7]~[#6]~[#6]~2~[#6]~[#6]~[#6]~[#6]~[#6]12" as the common SMARTS substructure. This is the fused ring of the benzodiazepine system but without bond type or atom aromaticity information. Use the first molecule as the reference depiction. All other depictions must have the depiction of their common substructure aligned to the reference.
The final image should be 400x400 pixels, which means each structure depiction should be 100x100 pixels. If the toolkit doesn't support multiple depictions in the same image then depict each to a different image. A desktop tool would need a way to write the image data to a GUI widget, but for now just write the result to a PNG or GIF file.
The point here is to show how to do the 2D structure alignment. The other details are less important. Use your own preferences for coloring scheme (eg, color on white background or on black background). You may assume the input structure has the correct MDL chemistry. (This is only relevant for toolkits which support multiple chemistry models.)
OpenEye/Python
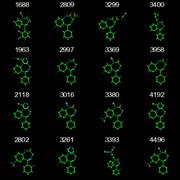
Output from OpenEye/Python
from openeye.oechem import *
from openeye.oedepict import *
CELL_WIDTH = CELL_HEIGHT = 100
def make_view():
view = OEDepictView(CELL_WIDTH, CELL_HEIGHT)
view.SetSuperAtoms(False)
view.SetAromaticCircles(False)
view.SetAromaticDashes(True)
view.SetColorOnBlack(True)
view.SetBackColor(0, 0, 0)
view.SetForeColor(255, 255, 255)
view.SetShowHydrogens(True)
view.SetDativeBonds(False)
return view
# This assumes the input was in proper SD format already
def prepare_mol_for_depiction(mol):
if mol.GetDimension() == 3:
OEPerceiveChiral(mol)
OE3DToBondStereo(mol)
OE3DToAtomStereo(mol)
OEAddDepictionHydrogens(mol)
core_subsearch = OESubSearch("[#7]~1~[#6]~[#6]~[#7]~[#6]~[#6]~2~[#6]~[#6]~[#6]~[#6]~[#6]12")
ifs = oemolistream("benzodiazepine.sdf.gz")
img = OE8BitImage(CELL_WIDTH*4, CELL_HEIGHT*4)
view = make_view()
at_first_structure = True
for record_num, mol in enumerate(ifs.GetOEGraphMols()):
prepare_mol_for_depiction(mol)
for match in core_subsearch.Match(mol):
if at_first_structure:
OEDepictCoordinates(mol)
# save the first structure's depiction coordinates
coords = [mol.GetCoords(atom) for atom in match.GetTargetAtoms()]
else:
# Align to the first structure's coordinates
fixed = [0] * mol.GetMaxAtomIdx()
for atom, coord in zip(match.GetTargetAtoms(), coords):
mol.SetCoords(atom, coord)
fixed[atom.GetIdx()] = 1
OEDepictFixedCoordinates(mol, fixed)
view.SetMolecule(mol)
view.RenderImage(img, at_first_structure,
record_num//4 * CELL_WIDTH, record_num %4 * CELL_HEIGHT)
at_first_structure = False
# Only align the first match
break
# Stop after 16 structures
if record_num == 15:
break
# Save the final image
ofs = oeofstream("align_oe.gif")
OEWriteGIF(ofs, img)