(tuned the parameters for nicer output) |
(Undo revision 4479 by 78.171.163.73 (talk)) Tag: rte-wysiwyg |
||
| (38 intermediate revisions by 15 users not shown) | |||
| Line 1: | Line 1: | ||
I don't know about you, but I have a hard time looking at anything other than trivial SMILES strings and SD files and understanding the contained molecular structure. Images help, a lot. Noel O'Boyle put together an incredible [http://baoilleach.webfactional.com/site_media/blog_b/depict.html molecular depiction comparison], so go there if you want to get a sense for the visual styles from different toolkits. |
I don't know about you, but I have a hard time looking at anything other than trivial SMILES strings and SD files and understanding the contained molecular structure. Images help, a lot. Noel O'Boyle put together an incredible [http://baoilleach.webfactional.com/site_media/blog_b/depict.html molecular depiction comparison], so go there if you want to get a sense for the visual styles from different toolkits. |
||
| − | The point here is to show |
+ | The point here is to show the details of how to take a structure without 2D coordinates, generate a layout, depict it, and save the results to a file. |
==Implementation== |
==Implementation== |
||
| Line 8: | Line 8: | ||
The depiction should go through all the steps in generating a standard depiction for any input SMILES, and should not take advantage of the simplicity of the test structure. |
The depiction should go through all the steps in generating a standard depiction for any input SMILES, and should not take advantage of the simplicity of the test structure. |
||
| + | |||
| + | ==CDK/Java== |
||
| + | |||
| + | [[File:Caffeine.png|thumb]] |
||
| + | |||
| + | CDK v1.5.12+ provides a new ''depict'' API simplifying image generation. An intermediate Depiction instance is generated that can then be written to raster (PNG,GIF,JPG) or vector (SVG,PDF) output. |
||
| + | <br /><br /><br /><br /><br /><br /><br /><br /><br /> |
||
| + | |||
| + | <source lang="java"> |
||
| + | import org.openscience.cdk.CDKConstants; |
||
| + | import org.openscience.cdk.interfaces.*; |
||
| + | import org.openscience.cdk.silent.SilentChemObjectBuilder; |
||
| + | import org.openscience.cdk.smiles.SmilesParser; |
||
| + | import java.awt.Color; |
||
| + | |||
| + | public class Main { |
||
| + | public static void main(String[] args) throws Exception { |
||
| + | IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance(); |
||
| + | SmilesParser smipar = new SmilesParser(bldr); |
||
| + | |||
| + | IAtomContainer mol = smipar.parseSmiles("CN1C=NC2=C1C(=O)N(C(=O)N2C)C"); |
||
| + | mol.setProperty(CDKConstants.TITLE, "caffeine"); |
||
| + | |||
| + | DepictionGenerator dptgen = new DepictionGenerator(); |
||
| + | // size in px (raster) or mm (vector) |
||
| + | // annotations are red by default |
||
| + | dptgen.withSize(200, 250) |
||
| + | .withMolTitle() |
||
| + | .withTitleColor(Color.DARK_GRAY); |
||
| + | dptgen.depict(mol) |
||
| + | .writeTo("~/caffeine.png"); |
||
| + | } |
||
| + | } |
||
| + | </source> |
||
| + | |||
| + | ==CDK/Groovy== |
||
| + | |||
| + | [[File:CTR2.png|thumb|alt=Caffeine depiction with the CDK toolkits.|Output from CDK/Groovy]] |
||
| + | |||
| + | When saves as depict.groovy and run with the following command, it will automatically download all required libraries: |
||
| + | |||
| + | |||
| + | |||
| + | groovy depict.groovy |
||
| + | |||
| + | <source lang="groovy"> |
||
| + | @GrabResolver( |
||
| + | name='idea', |
||
| + | root='http://ambit.uni-plovdiv.bg:8083/nexus/content/repositories/thirdparty/' |
||
| + | ) |
||
| + | @Grapes([ |
||
| + | @Grab( |
||
| + | group='org.openscience.cdk', |
||
| + | module='cdk-io', |
||
| + | version='1.4.11' |
||
| + | ), |
||
| + | @Grab( |
||
| + | group='org.openscience.cdk', |
||
| + | module='cdk-silent', |
||
| + | version='1.4.11' |
||
| + | ), |
||
| + | @Grab( |
||
| + | group='org.openscience.cdk', |
||
| + | module='cdk-sdg', |
||
| + | version='1.4.11' |
||
| + | ), |
||
| + | @Grab( |
||
| + | group='org.openscience.cdk', |
||
| + | module='cdk-smiles', |
||
| + | version='1.4.11' |
||
| + | ), |
||
| + | @Grab( |
||
| + | group='org.openscience.cdk', |
||
| + | module='cdk-renderawt', |
||
| + | version='1.4.11' |
||
| + | ) |
||
| + | ]) |
||
| + | |||
| + | import java.util.List; |
||
| + | import java.awt.*; |
||
| + | import java.awt.image.*; |
||
| + | import javax.imageio.*; |
||
| + | import org.openscience.cdk.silent.*; |
||
| + | import org.openscience.cdk.interfaces.*; |
||
| + | import org.openscience.cdk.layout.*; |
||
| + | import org.openscience.cdk.renderer.*; |
||
| + | import org.openscience.cdk.renderer.font.*; |
||
| + | import org.openscience.cdk.renderer.generators.*; |
||
| + | import org.openscience.cdk.renderer.visitor.*; |
||
| + | import org.openscience.cdk.smiles.SmilesParser; |
||
| + | import org.openscience.cdk.templates.*; |
||
| + | import org.openscience.cdk.renderer.generators.BasicSceneGenerator.Margin; |
||
| + | import org.openscience.cdk.renderer.generators.BasicSceneGenerator.ZoomFactor; |
||
| + | |||
| + | smiles = "CN1C=NC2=C1C(=O)N(C(=O)N2C)C" |
||
| + | int WIDTH = 200 |
||
| + | int HEIGHT = 250 |
||
| + | Rectangle drawArea = new Rectangle(WIDTH, HEIGHT); |
||
| + | Image image = new BufferedImage( |
||
| + | WIDTH, HEIGHT, BufferedImage.TYPE_INT_RGB |
||
| + | ); |
||
| + | smilesParser = new SmilesParser( |
||
| + | SilentChemObjectBuilder.getInstance() |
||
| + | ) |
||
| + | molecule = smilesParser.parseSmiles(smiles) |
||
| + | StructureDiagramGenerator sdg = |
||
| + | new StructureDiagramGenerator(); |
||
| + | sdg.setMolecule(molecule); |
||
| + | sdg.generateCoordinates(); |
||
| + | molecule = sdg.getMolecule(); |
||
| + | List generators = |
||
| + | new ArrayList(); |
||
| + | generators.add(new BasicSceneGenerator()); |
||
| + | generators.add(new BasicBondGenerator()); |
||
| + | generators.add(new BasicAtomGenerator()); |
||
| + | AtomContainerRenderer renderer = |
||
| + | new AtomContainerRenderer( |
||
| + | generators, new AWTFontManager() |
||
| + | ); |
||
| + | renderer.setup(molecule, drawArea); |
||
| + | model = renderer.getRenderer2DModel(); |
||
| + | model.set(ZoomFactor.class, (double)0.9); |
||
| + | Graphics2D g2 = (Graphics2D)image.getGraphics(); |
||
| + | g2.setColor(Color.WHITE); |
||
| + | g2.fillRect(0, 0, WIDTH, HEIGHT); |
||
| + | renderer.paint(molecule, new AWTDrawVisitor(g2)); |
||
| + | ImageIO.write( |
||
| + | (RenderedImage)image, "PNG", |
||
| + | new File("CTR2.png") |
||
| + | ); |
||
| + | </source> |
||
| + | |||
| + | ==OpenBabel/Rubabel== |
||
| + | <source lang="ruby"> |
||
| + | require 'rubabel' |
||
| + | mol = Rubabel["CCN1CCC(CC1)n2cc(CNc3cc(Cl)c4ncc(C#N)c(Nc5ccc(F)c(Cl)c5)c4c3)nn2"] |
||
| + | mol.title = 'Caffeine' |
||
| + | mol.write('caffeine.png') |
||
| + | </source> |
||
| + | |||
| + | ==Indigo/C== |
||
| + | [[Image:caffeine_indigo_2.png|thumb|right|alt=Caffeine depiction with the Indigo toolkit.|Output from Indigo]] |
||
| + | <source lang="c"> |
||
| + | #include "indigo.h" |
||
| + | #include "indigo-renderer.h" |
||
| + | |||
| + | int main (void) |
||
| + | { |
||
| + | int mol = indigoLoadMoleculeFromString("CN1C=NC2=C1C(=O)N(C(=O)N2C)C"); |
||
| + | indigoLayout(mol); /* if not called, will be done automatically by the renderer */ |
||
| + | indigoSetOption("render-output-format", "png"); |
||
| + | indigoSetOption("render-comment", "Caffeine"); |
||
| + | indigoSetOption("render-comment-position", "top"); |
||
| + | indigoSetOptionXY("render-image-size", 200, 250); |
||
| + | indigoSetOptionColor("render-background-color", 1.0, 1.0, 1.0); |
||
| + | indigoRenderToFile(mol, "caffeine.png"); |
||
| + | indigoFree(mol); /* if not called, will be freed automatically on exit */ |
||
| + | return 0; |
||
| + | } |
||
| + | </source> |
||
| + | ==Indigo/Java== |
||
| + | <source lang="java"> |
||
| + | package test; |
||
| + | import com.gga.indigo.*; |
||
| + | import java.io.*; |
||
| + | import java.util.*; |
||
| + | |||
| + | public class Main |
||
| + | { |
||
| + | public static void main (String[] args) throws java.io.IOException |
||
| + | { |
||
| + | Indigo indigo = new Indigo(); |
||
| + | IndigoRenderer renderer = new IndigoRenderer(indigo); |
||
| + | |||
| + | IndigoObject mol = indigo.loadMolecule("CN1C=NC2=C1C(=O)N(C(=O)N2C)C"); |
||
| + | mol.layout(); // if not called, will be done automatically by the renderer |
||
| ⚫ | |||
| + | indigo.setOption("render-comment", "Caffeine"); |
||
| + | indigo.setOption("render-comment-position", "top"); |
||
| + | indigo.setOption("render-image-size", 200, 250); |
||
| + | indigo.setOption("render-background-color", 1.0, 1.0, 1.0); |
||
| + | renderer.renderToFile(mol, "caffeine.png"); |
||
| + | } |
||
| + | } |
||
| + | </source> |
||
==Indigo/Python== |
==Indigo/Python== |
||
<source lang="python"> |
<source lang="python"> |
||
| + | from indigo import * |
||
| + | from indigo_renderer import * |
||
| + | |||
| + | indigo = Indigo() |
||
| + | renderer = IndigoRenderer(indigo) |
||
| + | |||
mol = indigo.loadMolecule("CN1C=NC2=C1C(=O)N(C(=O)N2C)C") |
mol = indigo.loadMolecule("CN1C=NC2=C1C(=O)N(C(=O)N2C)C") |
||
mol.layout() # if not called, will be done automatically by the renderer |
mol.layout() # if not called, will be done automatically by the renderer |
||
| Line 16: | Line 207: | ||
indigo.setOption("render-comment", "Caffeine") |
indigo.setOption("render-comment", "Caffeine") |
||
indigo.setOption("render-comment-position", "top") |
indigo.setOption("render-comment-position", "top") |
||
| − | indigo.setOption("render- |
+ | indigo.setOption("render-image-size", 200, 250) |
| ⚫ | |||
indigo.setOption("render-background-color", 1.0, 1.0, 1.0) |
indigo.setOption("render-background-color", 1.0, 1.0, 1.0) |
||
renderer.renderToFile(mol, "caffeine.png") |
renderer.renderToFile(mol, "caffeine.png") |
||
</source> |
</source> |
||
| + | |||
==OEChem/Python== |
==OEChem/Python== |
||
| Line 88: | Line 279: | ||
Draw.MolToFile(mol,"caffeine.png",size=(200,250)) |
Draw.MolToFile(mol,"caffeine.png",size=(200,250)) |
||
</source> |
</source> |
||
| + | |||
| + | ==Cactvs/Tcl== |
||
| + | |||
| + | [[Image:caffeine_cactvs.png|thumb|right|alt=Caffeine depiction with Cactvs.|Output from Cactvs/Tcl]] |
||
| + | |||
| + | <pre lang="tcl"> |
||
| + | prop setparam E_GIF width 200 height 250 bgcolor white \ |
||
| + | atomcolor black format png header Caffeine \ |
||
| + | filename caffeine.png |
||
| + | ens get "CN1C=NC2=C1C(=O)N(C(=O)N2C)C" E_GIF |
||
| + | </pre> |
||
| + | |||
| + | Btw, it is no accident that the rings are horizontally aligned, and the six-membered ring on the left. That's what a chemist expects, and quality depictions need to take this into account! |
||
| + | |||
| + | The white space around the molecule is intentional. There is a parameter (here with default value 8, a useful setting for typical drugs, but large for this rather small structure) which controls how many standard bonds should fit in the rendering. Smaller molecules are centered, larger ones shrunk dynamically to just fit into the drawing area. This avoids the dreaded "pumping bonds" display style where the bond length varies from image to image. |
||
| + | |||
| + | ==Cactvs/Python== |
||
| + | <pre lang="python"> |
||
| + | Prop.Setparam('E_GIF','width',200,'height',250,'bgcolor','white', |
||
| + | 'atomcolor','black','format','png','header','Caffeine','filename','caffeine.png') |
||
| + | Ens.Get('CN1C=NC2=C1C(=O)N(C(=O)N2C)C','E_GIF') |
||
| + | </pre> |
||
| + | |||
| + | The output is pixel-identical to the Tcl result. |
||
[[Category:SMILES]] |
[[Category:SMILES]] |
||
[[Category:depiction]] |
[[Category:depiction]] |
||
| Line 93: | Line 308: | ||
[[Category:RDKit/Python]] |
[[Category:RDKit/Python]] |
||
[[Category:Indigo/Python]] |
[[Category:Indigo/Python]] |
||
| + | [[Category:Indigo/C]] |
||
| + | [[Category:Indigo/Java]] |
||
| + | [[Category:Cactvs/Tcl]] |
||
| + | [[Category:CDK/Groovy]] |
||
| + | [[Category:CDK/Java]] |
||
Revision as of 10:55, 8 August 2016
I don't know about you, but I have a hard time looking at anything other than trivial SMILES strings and SD files and understanding the contained molecular structure. Images help, a lot. Noel O'Boyle put together an incredible molecular depiction comparison, so go there if you want to get a sense for the visual styles from different toolkits.
The point here is to show the details of how to take a structure without 2D coordinates, generate a layout, depict it, and save the results to a file.
Implementation
Depict the SMILES "CN1C=NC2=C1C(=O)N(C(=O)N2C)C" as an image of size 200x250 pixels. The image should be in PNG format if possible, otherwise in GIF format. If possible, give it the title "Caffeine". It should display the structure on a white background.
The depiction should go through all the steps in generating a standard depiction for any input SMILES, and should not take advantage of the simplicity of the test structure.
CDK/Java
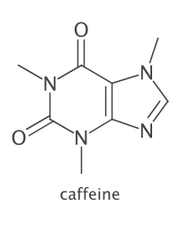
CDK v1.5.12+ provides a new depict API simplifying image generation. An intermediate Depiction instance is generated that can then be written to raster (PNG,GIF,JPG) or vector (SVG,PDF) output.
import org.openscience.cdk.CDKConstants;
import org.openscience.cdk.interfaces.*;
import org.openscience.cdk.silent.SilentChemObjectBuilder;
import org.openscience.cdk.smiles.SmilesParser;
import java.awt.Color;
public class Main {
public static void main(String[] args) throws Exception {
IChemObjectBuilder bldr = SilentChemObjectBuilder.getInstance();
SmilesParser smipar = new SmilesParser(bldr);
IAtomContainer mol = smipar.parseSmiles("CN1C=NC2=C1C(=O)N(C(=O)N2C)C");
mol.setProperty(CDKConstants.TITLE, "caffeine");
DepictionGenerator dptgen = new DepictionGenerator();
// size in px (raster) or mm (vector)
// annotations are red by default
dptgen.withSize(200, 250)
.withMolTitle()
.withTitleColor(Color.DARK_GRAY);
dptgen.depict(mol)
.writeTo("~/caffeine.png");
}
}
CDK/Groovy
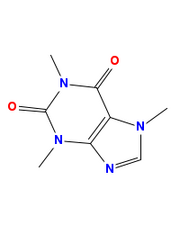
Output from CDK/Groovy
When saves as depict.groovy and run with the following command, it will automatically download all required libraries:
groovy depict.groovy
@GrabResolver(
name='idea',
root='http://ambit.uni-plovdiv.bg:8083/nexus/content/repositories/thirdparty/'
)
@Grapes([
@Grab(
group='org.openscience.cdk',
module='cdk-io',
version='1.4.11'
),
@Grab(
group='org.openscience.cdk',
module='cdk-silent',
version='1.4.11'
),
@Grab(
group='org.openscience.cdk',
module='cdk-sdg',
version='1.4.11'
),
@Grab(
group='org.openscience.cdk',
module='cdk-smiles',
version='1.4.11'
),
@Grab(
group='org.openscience.cdk',
module='cdk-renderawt',
version='1.4.11'
)
])
import java.util.List;
import java.awt.*;
import java.awt.image.*;
import javax.imageio.*;
import org.openscience.cdk.silent.*;
import org.openscience.cdk.interfaces.*;
import org.openscience.cdk.layout.*;
import org.openscience.cdk.renderer.*;
import org.openscience.cdk.renderer.font.*;
import org.openscience.cdk.renderer.generators.*;
import org.openscience.cdk.renderer.visitor.*;
import org.openscience.cdk.smiles.SmilesParser;
import org.openscience.cdk.templates.*;
import org.openscience.cdk.renderer.generators.BasicSceneGenerator.Margin;
import org.openscience.cdk.renderer.generators.BasicSceneGenerator.ZoomFactor;
smiles = "CN1C=NC2=C1C(=O)N(C(=O)N2C)C"
int WIDTH = 200
int HEIGHT = 250
Rectangle drawArea = new Rectangle(WIDTH, HEIGHT);
Image image = new BufferedImage(
WIDTH, HEIGHT, BufferedImage.TYPE_INT_RGB
);
smilesParser = new SmilesParser(
SilentChemObjectBuilder.getInstance()
)
molecule = smilesParser.parseSmiles(smiles)
StructureDiagramGenerator sdg =
new StructureDiagramGenerator();
sdg.setMolecule(molecule);
sdg.generateCoordinates();
molecule = sdg.getMolecule();
List generators =
new ArrayList();
generators.add(new BasicSceneGenerator());
generators.add(new BasicBondGenerator());
generators.add(new BasicAtomGenerator());
AtomContainerRenderer renderer =
new AtomContainerRenderer(
generators, new AWTFontManager()
);
renderer.setup(molecule, drawArea);
model = renderer.getRenderer2DModel();
model.set(ZoomFactor.class, (double)0.9);
Graphics2D g2 = (Graphics2D)image.getGraphics();
g2.setColor(Color.WHITE);
g2.fillRect(0, 0, WIDTH, HEIGHT);
renderer.paint(molecule, new AWTDrawVisitor(g2));
ImageIO.write(
(RenderedImage)image, "PNG",
new File("CTR2.png")
);
OpenBabel/Rubabel
require 'rubabel'
mol = Rubabel["CCN1CCC(CC1)n2cc(CNc3cc(Cl)c4ncc(C#N)c(Nc5ccc(F)c(Cl)c5)c4c3)nn2"]
mol.title = 'Caffeine'
mol.write('caffeine.png')
Indigo/C
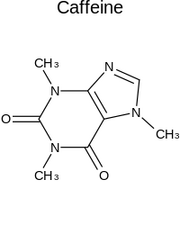
Output from Indigo
#include "indigo.h"
#include "indigo-renderer.h"
int main (void)
{
int mol = indigoLoadMoleculeFromString("CN1C=NC2=C1C(=O)N(C(=O)N2C)C");
indigoLayout(mol); /* if not called, will be done automatically by the renderer */
indigoSetOption("render-output-format", "png");
indigoSetOption("render-comment", "Caffeine");
indigoSetOption("render-comment-position", "top");
indigoSetOptionXY("render-image-size", 200, 250);
indigoSetOptionColor("render-background-color", 1.0, 1.0, 1.0);
indigoRenderToFile(mol, "caffeine.png");
indigoFree(mol); /* if not called, will be freed automatically on exit */
return 0;
}
Indigo/Java
package test;
import com.gga.indigo.*;
import java.io.*;
import java.util.*;
public class Main
{
public static void main (String[] args) throws java.io.IOException
{
Indigo indigo = new Indigo();
IndigoRenderer renderer = new IndigoRenderer(indigo);
IndigoObject mol = indigo.loadMolecule("CN1C=NC2=C1C(=O)N(C(=O)N2C)C");
mol.layout(); // if not called, will be done automatically by the renderer
indigo.setOption("render-output-format", "png");
indigo.setOption("render-comment", "Caffeine");
indigo.setOption("render-comment-position", "top");
indigo.setOption("render-image-size", 200, 250);
indigo.setOption("render-background-color", 1.0, 1.0, 1.0);
renderer.renderToFile(mol, "caffeine.png");
}
}
Indigo/Python
from indigo import *
from indigo_renderer import *
indigo = Indigo()
renderer = IndigoRenderer(indigo)
mol = indigo.loadMolecule("CN1C=NC2=C1C(=O)N(C(=O)N2C)C")
mol.layout() # if not called, will be done automatically by the renderer
indigo.setOption("render-output-format", "png")
indigo.setOption("render-comment", "Caffeine")
indigo.setOption("render-comment-position", "top")
indigo.setOption("render-image-size", 200, 250)
indigo.setOption("render-background-color", 1.0, 1.0, 1.0)
renderer.renderToFile(mol, "caffeine.png")
OEChem/Python
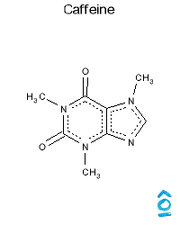
Output from OpenEye/Python
from openeye.oechem import *
from openeye.oedepict import *
smiles = "CN1C=NC2=C1C(=O)N(C(=O)N2C)C"
mol = OEMol()
OEParseSmiles(mol, smiles)
mol.SetTitle("Caffeine")
OEAssignAromaticFlags(mol)
OESetDimensionFromCoords(mol)
OESuppressHydrogens(mol)
OEAddDepictionHydrogens(mol)
OEDepictCoordinates(mol)
OEMDLPerceiveBondStereo(mol)
view = OEDepictView(200, 250)
view.SetSuperAtoms(False)
view.SetAromaticCircles(False)
view.SetAromaticDashes(True)
# Make this a black and white depiction
view.SetColorOnBlack(False)
view.SetBackColor(255, 255, 255)
view.SetForeColor(0, 0, 0)
view.SetShowHydrogens(True)
view.SetDativeBonds(False)
#view.SetLogo(False)
img = OE8BitImage(view.XRange(), view.YRange())
view.SetMolecule(mol)
view.RenderImage(img, True, 0, 0)
# OpenEye does not have a PNG writer. See
# http://www.dalkescientific.com/writings/diary/archive/2007/05/23/oe8bitimage_to_png.html
# for one way to export the image data to PIL, which can generate the PNG.
# For C++ programmers, see toolkits/examples/ogham/mol2png.cpp
ofs = oeofstream("caffeine.gif")
OEWriteGIF(ofs, img)
RDKit/Python
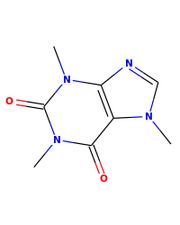
Output from RDKit/Python
from rdkit.Chem import AllChem
from rdkit.Chem import Draw
smiles = "CN1C=NC2=C1C(=O)N(C(=O)N2C)C"
mol = AllChem.MolFromSmiles(smiles)
# technically this step isn't required since the drawing code
# will automatically add a 2D conformation to a molecule that has
# no conformation information, I'm including it to show how to
# generate 2D coords with the RDKit:
AllChem.Compute2DCoords(mol)
Draw.MolToFile(mol,"caffeine.png",size=(200,250))
Cactvs/Tcl
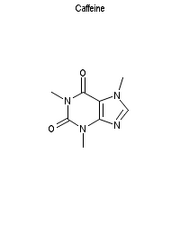
Output from Cactvs/Tcl
prop setparam E_GIF width 200 height 250 bgcolor white \
atomcolor black format png header Caffeine \
filename caffeine.png
ens get "CN1C=NC2=C1C(=O)N(C(=O)N2C)C" E_GIF
Btw, it is no accident that the rings are horizontally aligned, and the six-membered ring on the left. That's what a chemist expects, and quality depictions need to take this into account!
The white space around the molecule is intentional. There is a parameter (here with default value 8, a useful setting for typical drugs, but large for this rather small structure) which controls how many standard bonds should fit in the rendering. Smaller molecules are centered, larger ones shrunk dynamically to just fit into the drawing area. This avoids the dreaded "pumping bonds" display style where the bond length varies from image to image.
Cactvs/Python
Prop.Setparam('E_GIF','width',200,'height',250,'bgcolor','white',
'atomcolor','black','format','png','header','Caffeine','filename','caffeine.png')
Ens.Get('CN1C=NC2=C1C(=O)N(C(=O)N2C)C','E_GIF')
The output is pixel-identical to the Tcl result.