Greglandrum (talk | contribs) (Adding categories) |
Greglandrum (talk | contribs) No edit summary |
||
| Line 73: | Line 73: | ||
ofs = oeofstream("highlight_oe.gif") |
ofs = oeofstream("highlight_oe.gif") |
||
OEWriteGIF(ofs, img) |
OEWriteGIF(ofs, img) |
||
| + | </source> |
||
| + | |||
| + | ==RDKit/Python== |
||
| + | [[Image:3016_highlighted.rdkit.png|thumb|right|alt=Mol 3016 depiction with the RDKit toolkit.|Output from RDKit/Python]] |
||
| + | |||
| + | <source lang="python"> |
||
| + | from rdkit import Chem |
||
| + | suppl = Chem.SDMolSupplier('benzodiazepine.sdf') |
||
| + | tgt=None |
||
| + | for mol in suppl: |
||
| + | if not mol: continue |
||
| + | if mol.GetProp('_Name')=='3016': |
||
| + | tgt=mol |
||
| + | break |
||
| + | |||
| + | patt = Chem.MolFromSmarts("c1ccc2c(c1)C(=NCCN2)c3ccccc3") |
||
| + | # get the matching atoms: |
||
| + | matching = mol.GetSubstructMatch(patt) |
||
| + | |||
| + | from rdkit.Chem import Draw |
||
| + | |||
| + | # By default the RDKit colors atoms by element in depictions. |
||
| + | # We can turn this off by replacing the element dictionary |
||
| + | # in MolDrawing: |
||
| + | from rdkit.Chem.Draw import MolDrawing |
||
| + | from collections import defaultdict |
||
| + | MolDrawing.elemDict=defaultdict(lambda : (0,0,0)) |
||
| + | |||
| + | Draw.MolToFile(tgt,'3016_highlighted.rdkit.png',size=(200,250),highlightAtoms=matching ) |
||
| + | |||
</source> |
</source> |
||
[[Category:depiction]] |
[[Category:depiction]] |
||
Revision as of 05:50, 1 March 2010
The benzodiazepine data set was generated by doing a PubChem search for the SMARTS query "C2(CN=C(C1=CC(=CC=C1N2[*])[*])C3=CC=CC=C3[*])=[*]" where the [*] are for the R-groups. The query pattern was generated by the PubChem sketcher, and the aromatic form without R-groups is "c1ccc2c(c1)C(=NCCN2)c3ccccc3".
This task will extract a molecule from that data set and depict it with part of the substructure highlighted.
Implementation
Read record 3016 from the benzodiazepine SD file. Find all atoms which match the SMARTS "c1ccc2c(c1)C(=NCCN2)c3ccccc3" and highlight them in red. All other atoms must be drawn in black.
The resulting image should be 200x250 pixels and on a white background. The resulting image file should be in PNG (preferred) or GIF format.
OpenEye/Python
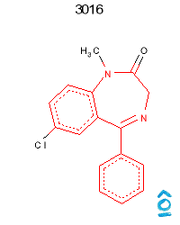
Output from OpenEye/Python
from openeye.oechem import *
from openeye.oedepict import *
# Find the structure matching the given title
def get_molecule_by_title(ifs, title):
for mol in ifs.GetOEGraphMols():
if mol.GetTitle() == title:
return mol
raise KeyError(title)
def do_layout_and_make_view(mol):
# These operations may modify the structure
OEAssignAromaticFlags(mol)
OESetDimensionFromCoords(mol)
OESuppressHydrogens(mol)
OEAddDepictionHydrogens(mol)
OEDepictCoordinates(mol)
OEMDLPerceiveBondStereo(mol)
view = OEDepictView(200, 250)
view.SetSuperAtoms(False)
view.SetAromaticCircles(False)
view.SetAromaticDashes(True)
view.SetColorOnBlack(False)
view.SetBackColor(255, 255, 255)
view.SetForeColor(0, 0, 0)
view.SetShowHydrogens(True)
view.SetDativeBonds(False)
view.SetMolecule(mol)
return view
# Set the color of each atom, as specified
def color_atoms(view, atoms, (r, g, b)):
for atom in atoms:
astyle = view.AStyle(atom.GetIdx())
astyle.r, astyle.g, astyle.b = r, g, b
# Get the right molecule
ifs = oemolistream("benzodiazepine.sdf.gz")
mol = get_molecule_by_title(ifs, "3016")
# prepare it for depiction and make everything black
view = do_layout_and_make_view(mol)
color_atoms(view, mol.GetAtoms(), (0, 0, 0))
# Define the core pattern and make all matches red
core_subsearch = OESubSearch("c1ccc2c(c1)C(=NCCN2)c3ccccc3")
for match in core_subsearch.Match(mol, True):
color_atoms(view, match.GetTargetAtoms(), (255, 0, 0))
# Render to an image and write the result as a GIF
img = OE8BitImage(view.XRange(), view.YRange())
view.RenderImage(img, True, 0, 0)
ofs = oeofstream("highlight_oe.gif")
OEWriteGIF(ofs, img)
RDKit/Python
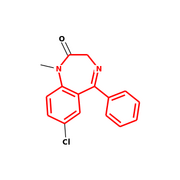
Output from RDKit/Python
from rdkit import Chem
suppl = Chem.SDMolSupplier('benzodiazepine.sdf')
tgt=None
for mol in suppl:
if not mol: continue
if mol.GetProp('_Name')=='3016':
tgt=mol
break
patt = Chem.MolFromSmarts("c1ccc2c(c1)C(=NCCN2)c3ccccc3")
# get the matching atoms:
matching = mol.GetSubstructMatch(patt)
from rdkit.Chem import Draw
# By default the RDKit colors atoms by element in depictions.
# We can turn this off by replacing the element dictionary
# in MolDrawing:
from rdkit.Chem.Draw import MolDrawing
from collections import defaultdict
MolDrawing.elemDict=defaultdict(lambda : (0,0,0))
Draw.MolToFile(tgt,'3016_highlighted.rdkit.png',size=(200,250),highlightAtoms=matching )